Avicenna Journal of Medical Biochemistry. 10(1):52-57.
doi: 10.34172/ajmb.2022.07
Original Article
Screening of Streptococcus mutans Sortase A Via Myricetin-Like Inhibitors: In Vitro Evaluation and Molecular Docking-Based Virtual
Mona Maghsoodlou 1  , Leila Fozouni 1, *
, Leila Fozouni 1, *  , Ali Salehnia Sammak 2
, Ali Salehnia Sammak 2 
Author information:
1Department of Microbiology, Gorgan Branch, Islamic Azad University, Gorgan, Iran
2Department of Microbiology, Rasht Branch, Islamic Azad University, Rasht, Iran
*
Corresponding author: Leila Fozouni, Shahid Kalantari´s Boulevard Daneshgah Street, Department of Microbiology, Gorgan, Iran. Tel:+989111518674 Email:
L.fozouni@gorganiau.ac.ir
Abstract
Background: Dental caries is one of the most common causes threatening human health globally. Sortase A (Srt A) as a transpeptidase, mediates the attachment of the Streptococcus mutans cell wall to dental surfaces by biofilm formation. Due to the development of multidrug-resistance bacteria, attempting to discover growth inhibitors is logical and promising.
Objectives: The current study aimed at the experimental and docking-based virtual screening of myricetinlike inhibitors for the inhibition of Srt A enzyme in S. mutans isolates.
Methods: Sixty-three S. mutans were isolated from pupils based on cultural, morphological, and biochemical characteristics (N=150). After identifying the srtA gene using the polymerase chain reaction (PCR) with specific primers, a broth microdilution test was conducted according to CLSI-2020 criteria to determine the minimum inhibitory concentration (MIC) of myricetin. The in silico exploration of Srt A inhibitors was performed using AutoDock 4.2.6.
Results: The frequency of S. mutans isolates containing the srtA gene was 87.3% of which, fifty isolates (79.4%) were categorized as susceptible to myricetin (MIC,≤16 μg/mL). Of 20 ligands having a high degree of similarity with myricetin, the best docking results were related to ligand 2.
Conclusion: It was concluded that myricetin has an inhibitory effect on oral bacteria in vitro, and ligand 2 had the most negative binding energy (-4.66 kcal/mol) and favorably interacts with the key amino acid residues at the active site of Srt A. Accordingly, this ligand can be utilized as a lead compound for further studies to discover novel inhibitors targeting Srt A in S. mutans.
Keywords: Streptococcus mutans, Molecular docking, Myricetin, Sortase A,
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Maghsoodlou M, Fozouni L, Salehnia Sammak A. Screening of streptococcus mutans sortase a via myricetin-like inhibitors: in vitro evaluation and molecular docking-based virtual. Avicenna J Med Biochem. 2022; 10(1):52-57. doi:10.34172/ ajmb.2022.07
Background
Tooth infection such as dental caries has continuously become the main public health concern globally. Tooth decay contains biological, environmental, and lifestyle-related risk factors. Given biological agents, oral microflora specified in both the root and crown of tooth possess an important situation in dental health (1,2). The main pathogen of the root and coronal caries are viridans-group Streptococci, especially Streptococcus mutans. S. mutans uses the sortase A (Srt A) enzyme for anchoring several surface proteins to the cell walls and forms a biofilm on the tooth surface (3,4). The biofilm production, the so-called dental plaque, is one of the significant virulence features of S. mutans that adheres bacteria to the surface of the teeth. The tooth continuously displays enamel decalcification, ultimately resulting in its decay because of the biofilm generation and the acid-manufacturing capacity of carbohydrate fermentation by bacteria. Several studies have reported that the presence of Streptococcus spp. reduces in the cell wall after the removal of Srt A; therefore, this enzyme possesses a key function in the interactions between Streptococcus spp. and their host and is concerned as a promising target for the treatment of tooth caries (5-7). To date, some Srt A inhibitors have been discovered that are mostly derived from flavonoid compounds, including myricetin (8). However, only a few small molecules are available for inhibiting the transpeptidase by myricetin, which is a naturally benzo-α-pyrone flavonoid derivative. Antibiotics utilized in dentistry were found to raise bacterial resistance, thus naturally occurring ingredients can make a favorable contribution to treating dental caries. Moreover, the computer-based drug design has appeared as a popular procedure for conducting high-throughput virtual screening of potential medicinal compounds, therefore decreasing time and experimental validation (9,10). In an attempt, the researchers exerted a high-throughput virtual screening procedure to recognize novel potential Srt A suppressors in S. mutans. They identified the potential suppressors to hinder the Srt A catalysis (11). Similarly, other researchers showed that the curcumin analogs can bind to the active site of the Srt A enzyme in a stable conformation. They suggested that the analog CA51 is a promising inhibitor of Srt A and can be utilized as a new antimicrobial ligand against Enterococcus faecalis infection (12). Considering that finding the inhibitors and their analogs against the Srt A enzyme in S. mutans has a key role in controlling dental caries, this study aimed to screen the myricetin-like inhibitors for Srt A in S. mutans to find a potential inhibitor that targets this enzyme.
Materials and Methods
Experimental Characteristics and Study Population
In this experimental study, 150 samples were taken from the buccal and lingual surfaces of the posterior teeth of elementary school children aged 7-12 years (with the same sex ratio) in Gorgan, northern Iran.The students were excluded from the study if they had used antibiotics within the past three months or had systemic or immunodeficiency disorders. The wooden toothpicks samples were collected and transferred to the laboratory in sterile tubes containing the brain heart infusion broth (Merck, Germany) and buffer. After two hours of incubation at 37°C, the samples were homogenized by vortexing and plated on Mitis Salivarius Bacitracin agar (HiMedia, India) for 24 hours at 37°C in anaerobic conditions. The S. mutans isolates were identified based on the colony morphology, Gram-staining, and biochemical tests (hemolysis, catalase, bile esculin, optochin susceptibility, methyl red/Voges-Proskauer, and arginine dihydrolase), as well as the fermentation of mannitol, lactose, salicin, and trehalose (13). Then, the broth microdilution test was applied according to CLSI M100 (2020) criteria to determine the minimum inhibitory concentration (MIC) of myricetin (14). First, a suspension (1.5 × 108 CFU/mL) of the 24-hour bacterial cultures in Müller Hinton Broth (Merck, Germany) was prepared spectrophotometrically. After preparing myricetin suspension in 1% of dimethyl sulfoxide, the concentration range of myricetin stock (Sigma Aldrich, USA) was determined at 512 -1 µg/mL. After overnight incubation at 37 °C in anaerobic conditions, the growth rate was measured and compared with that of the positive (without the myricetin) and negative (without bacterial suspension) control. The minimum concentration inhibiting bacterial growth up to 50% or 90%, compared to positive controls, is considered MIC50 and MIC90, respectively. S. mutans PTCC35668 was used as the control. The DNA of S. mutans isolates was extracted by the glass bead/phenol-chloroform method. DNA was employed as a template for srtA gene amplification using specific primers ‘F: 5’- CTCGGATCCAAACCTCATAT TGATAGTTATTTACATGAC-3, R: 5’- CTCGGTACCTTA TTTAATCTGTTCTGCTATAAATATTTTACGC -3’ taken from Gene Bank No. AF162657 (15,16). Amplification was programmed for an initial denaturation at 94°C for 1 minute, followed by 31 cycles of denaturation at 92°C for 1 minute, annealing at 55°C for 2 minutes, extension at 72°C for 3 minutes, and a final extension at 75°C for 5 minutes. Polymerase chain reaction (PCR) products were electrophoresed on 1% agarose gel, and the bands were visualized under UV light after staining with ethidium bromide. After verifying the normality of data distribution using the Kolmogorov-Smirnov test, quantitative and qualitative data were analyzed by an independent t test and chi-square test, respectively.
Docking for Potential Inhibitors of S. mutans Srt A
The appropriate crystallographic structure of the S. mutans Srt A enzyme containing the central catalytic site was selected from the PDB database with the ID 4TQX. The PubChem database and ChemSketch 12.0 molecular modeling program were used to obtain the chemical structure of myricetin-like molecules and create the structure of the molecules, respectively. The HyperChem 8.0 molecular modeling and The PyRx 0.8 virtual screening software were prepared to optimize molecules in terms of energy and evaluation of molecules. After obtaining the results, the binding affinity of all conformers related to each molecule was examined to select the best candidates for docking using AutoDock 4.2.6 molecular modeling simulation software. Electrostatic interactions for each protein atom were investigated by Autogrid 4.0.0, a program, which pre-calculates the grid map of interaction energy for different atom types. The box size 42*42*44 and point spacing 0.375 (equivalent to a quarter of the carbon-carbon bond length) were applied for inhibitor docking. The center of the two copper ions in the active site was designated as the center of the grid box. According to the calculation of the free energy of ligand binding, some ligands were selected for further docking. At this stage, after preparing the protein and its ligands in PDBQT format, the areas where the ligands interact with the protein were identified through the formation of a Grid Box. The best conformer for docking was selected using the relevant clustering diagram and physical parameters such as binding energy, inhibitory constant, and ligand efficiency. In other words, first, the best cluster was obtained according to the number of conformers in each cluster and the binding energy. Then, the best conformer was selected from each cluster by the three above-mentioned physical parameters. If the value of each physical parameter in each cluster was the lowest for a conformer, that conformer was considered as the final docking choice against Srt A. Finally, the interaction of myricetin and its analogs was evaluated and compared in more detail against the Srt A binding site via the Ligplot 4.5.3 computer program, which creates a schematic 2D representation of the protein-ligand complex (12).
Results
Results of In Vitro Assay
Streptococcus mutans (63, 42%) was the predominant streptococcal species. Other isolated species included Streptococcus sanguinis (28.11%) and Streptococcus oralis (8.47%). The presence of 348-bp fragments compared with the DNA marker indicated the presence of the srtA gene and confirmed the accuracy of the reaction. The results revealed that the frequency of S. mutans isolates containing srtA was 87.3% of which 79.4% (MIC, ≤ 16 μg/mL) of the isolates were categorized as susceptible to myricetin.The comparative in vitro susceptibilities of the S. mutans isolates to myricetin showed that the MIC50 was 128 µg/mL in comparison with the range of the reference strain (MIC = 32 µg/mL, P = 0.37) and was 4-fold lower than the MIC90 value (512 µg/mL) against the S. mutans isolate (Figure 1). The changes to the MIC of myricetin demonstrated that the difference between the growth and lack of growth of S. mutansisolates containing srtA was significant (P = 0.01).

Figure 1.
Minimum Inhibitory Concentrations of Myricetin Against Streptococcus mutans Isolates.
.
Minimum Inhibitory Concentrations of Myricetin Against Streptococcus mutans Isolates.
Data of Docking and Model Validation
In the present study, according to the binding site of the myricetin inhibitor in the Srt A enzyme and its inhibitory mechanism, first, a total of 20 ligands having a high degree of similarity with the myricetin molecule were selected and then analyzed via AutoDock (Figure 2). The matching between the structure of Srt A with myricetin and the ligands represented that the location of the active site is similar. The ligands were docked against Srt A for confirming the validity of the active site. The findings revealed that the key residues phe143 and thr146 were localized in the anticipated active center, and two or three hydrogen bonds between Srt A and its ligands were generated accordingly. This result demonstrated that the determined active site in the Srt A enzyme via AutoDock is comparatively precise and can be used for further virtual screening. The testing sets including 5 positive and 15 negative ligands were applied to measure the accuracy of AutoDock for screening Srt A inhibitors. The 50% of top-ranked ligands were docked via the Hawkins GB/SA score, accounted for 0.95 of the highest area under the receiver operating characteristic curve (data not shown). After studying the pharmacokinetic and quasi-pharmacological properties of the ligands, a total of 4 ligands having a high affinity for Srt A protein were selected as the candidate inhibitors. Each ligand was individually placed in the active site of the enzyme, and the amount of binding affinity, the number of hydrogen bonds with each amino acid, and the inhibitory power were obtained accordingly. The myricetin has a hydrogen bond with the amino acid phe143, which is 2.92 A° in length, and has a hydrophobic bond with 7 amino acids. Ligands 2 and 16 had three hydrogen bonds at the active site of the enzyme, namely, two hydrogen bonds with phe143 and one hydrogen bond with thr146. These ligands were also hydrophobically linked to 7 amino acids. Ligands 12 and 14 had two hydrogen bonds with he143, along with a total of 7 hydrophobic bonds (Figure 3).
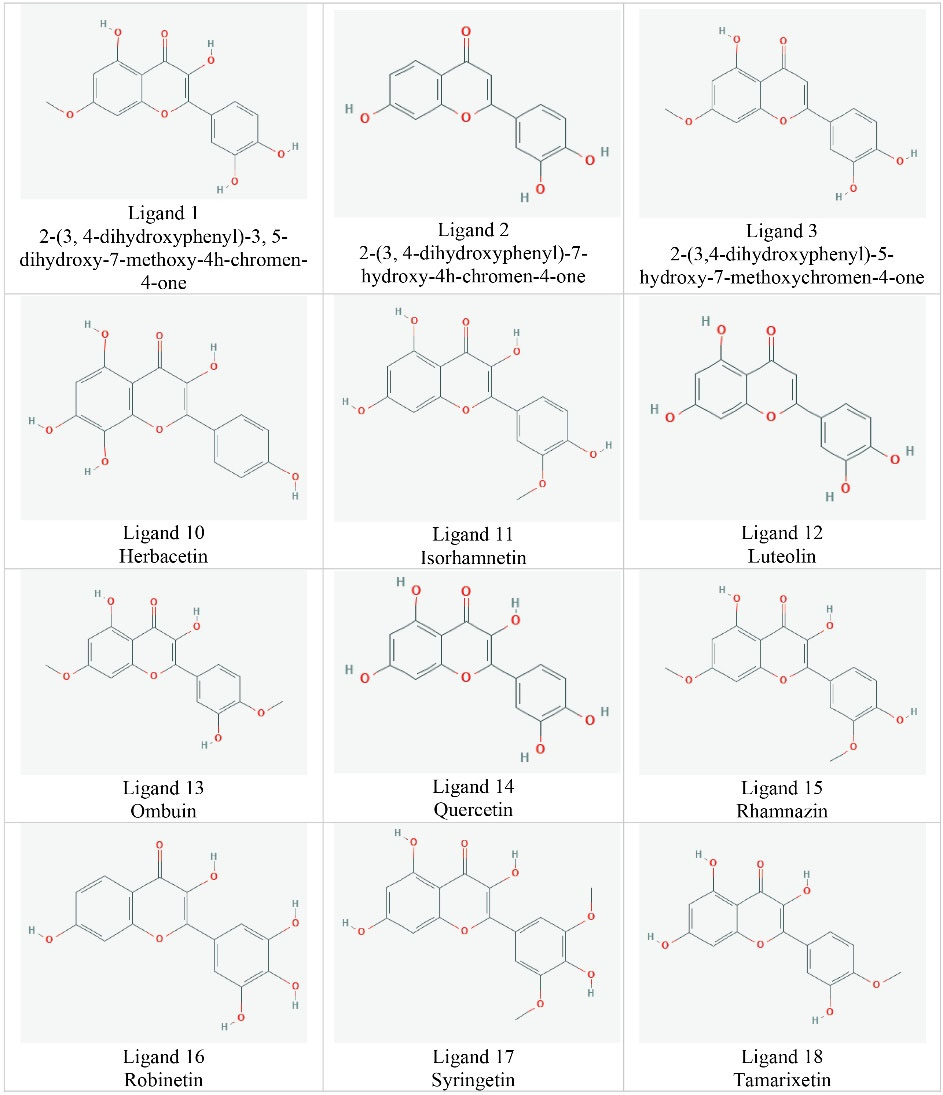
Figure 2.
The Structure of Ligands Having a High Degree of Similarity With Myricetin Molecule.
.
The Structure of Ligands Having a High Degree of Similarity With Myricetin Molecule.
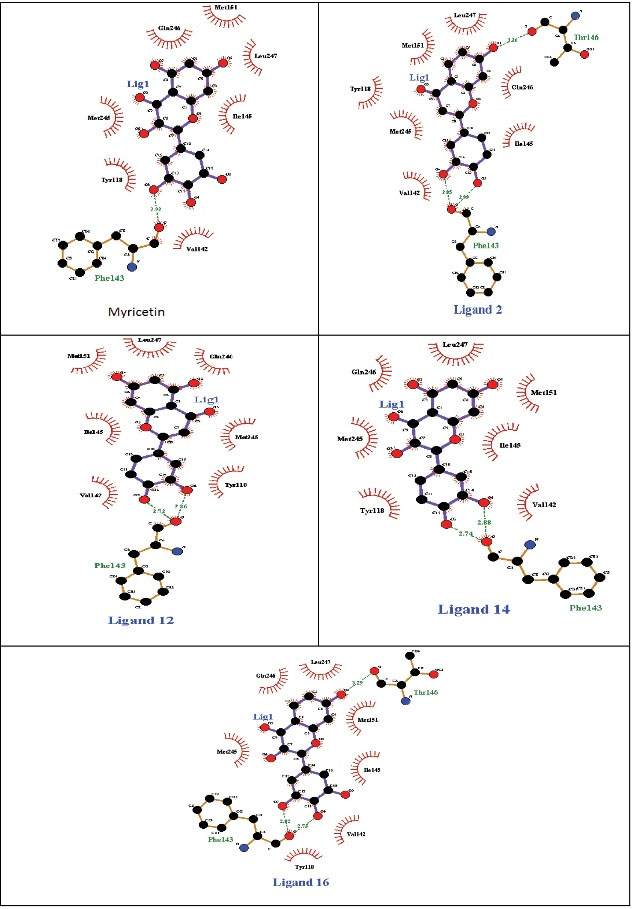
Figure 3.
Interaction of Myricetin and Ligands 2, 12, 14, and 16 With the Binding Site of the Sortase A Enzyme.
.
Interaction of Myricetin and Ligands 2, 12, 14, and 16 With the Binding Site of the Sortase A Enzyme.
The main molecule of myricetin has binding energy and an inhibitory constant of -4.19 kcal mol−1 and 851.28, respectively. Ligand 2 had the lowest inhibitory constant and binding energy among other ligands. Moreover, this ligand had lower binding energy and higher inhibitory efficacy than those of myricetin (Table 1). The most potential ligands displayed more favorable binding energy in contrast to myricetin at -4.19 kcal mol-1. Albeit myricetin belonged to flavonoid compounds, the binding energy was the lowest compared with other ligands.
Table 1.
The Value of Physical Parameters Associated With Each Ligand Targeting Sortase A
|
Ligand Name
|
Myricetin
|
Ligand 2
|
Ligand 12
|
Ligand 14
|
Ligand 16
|
| Binding energy (kcal mol−1) |
-4.19 |
-4.66 |
-4.3 |
4.46 |
-4.33 |
| Ligand efficiency (kcal mol−1) |
0.18 |
0.23 |
0.2 |
0.2 |
0.2 |
| Inhibition constant (ki) |
851.28 |
382.64 |
699.87 |
534.23 |
673.22 |
Note. Ligand 2: 2-(3,4-dihydroxyphenyl)-7-hydroxy-4H-chromen-4-one; Ligand 12: Luteolin; Ligand 14: Quercetin; Ligand 16: Robinetin.
Discussion
According to the World Health Organization, the problem of tooth decay remains worldwide, especially in socially disadvantaged groups (17). The tooth decay caused by drug-resistant oral bacteria has determined the need to discover new ways to combat these bacteria. Some researchers have confirmed the high role of Srt A in the formation of bacterial biofilms by modulating anchor surface proteins to the cell wall and eventually tooth decay (18,19). In the present study, more than 80% of the isolates contained the srtA gene. Researchers in Canada (20) and Iran (19) observed that the inactivation of the srtA gene reduces biofilm formation. Therefore, according to widespread reports of drug resistance (13,17), controlling it with natural compounds and enzymatic inhibitors seems reasonable. In this study, the changes to the MIC of myricetin showed that there was a significant difference between the growth and lack of growth of S. mutans isolates containing srtA. Progress in silico procedures has enabled virtual screening to have a promising effect on drug discovery. Docking-based virtual screening is based on predicting the affinity of each compound in the dataset via docking to an X-ray crystallographic structure. Docking also evaluates the binding mode of compounds with the amino acids present in the active pocket of the intended protein/enzyme (21,22). The results of the current study indicated the substantial applicability and accuracy of molecular docking. Similarly, this validation method was exerted by Luo et al (11) and Sivaramakrishnan et al (12) for in silico identification of inhibitors targeting sortase. Similar studies have been recorded considering the recognition of the Srt A inhibitors of Streptococcus spp. As well (23-25). However, the exact mechanism behind this inhibition is not yet clear. A variety of plant natural ingredients, especially flavonoid compounds, can hinder sortase. Additionally, the branched chains of flavone can reduce Srt A activity. (26). Some researchers observed that the carbonyl group can have an important function in the mechanism of Srt A suppression (11,26). Some inhibitors such as morin and curcumin can decrease the activity of Srt A via interacting with key cysteine that takes part in breaking the bond T-G from the motif LPXTG in the enzyme (27-29). Overall, it seems that the ligand targeting Srt A in this study may have occupied the binding site of the motif LPXTG to decrease transpeptidation and thereby affect biofilm production. Considering that myricetin showed a good inhibitory effect at high concentrations in vitro, docking-based virtual screening opens a promising window toward the identification of ligands having a higher inhibitory potential than chemical and natural compounds. Likewise, other researchers found that molecular docking can identify ligands having a higher inhibitory potential against S. mutans Srt A when compared to natural ingredients (30,31).
Conclusion
According to the results of the present study, myricetin exhibited antibacterial activity and inhibitory effects against S. mutansisolates in vitro. Docking-based virtual screening in this research led to the identification of a promising ligand against S. mutans Srt A. This ligand was the best compound for the selective inhibitor because it favorably interacts with important amino acids located in the active site of Srt A. However, further in vitro and in vivo studies are required to determine the Srt A inhibitory activity of the ligand identified in this research.
Acknowledgements
This article was extracted from an MSc thesis conducted by Mona Maghsoodlou and supported by the Research Council of the Islamic Azad University of Gorgan, Iran. The authors are grateful to all those who assisted us in this study.
Authors’ Contribution
LF contributed to the study concept and edited the final manuscript. MM performed laboratory examinations and interpreted the data. All authors discussed the results and implications and provided their comments during all stages.
Conflict of Interests
The authors declare that there is no conflict of interests.
Ethical Issues
The study was performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of I.A.U, Aliabad Branch (code No.: 1398.006), Golestan, Iran.
Funding/Support
No financial interests related to the content of this manuscript are declared.
References
- Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C. The global burden of oral diseases and risks to oral health. Bull World Health Organ 2005; 83(9):661-9. [ Google Scholar]
- Selwitz RH, Ismail AI, Pitts NB. Dental caries. Lancet 2007; 369(9555):51-9. doi: 10.1016/s0140-6736(07)60031-2 [Crossref] [ Google Scholar]
- Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev 2002; 15(4):613-30. doi: 10.1128/cmr.15.4.613-630.2002 [Crossref] [ Google Scholar]
- Doern CD, Burnham CA. It’s not easy being green: the viridans group streptococci, with a focus on pediatric clinical manifestations. J Clin Microbiol 2010; 48(11):3829-35. doi: 10.1128/jcm.01563-10 [Crossref] [ Google Scholar]
- Clancy KW, Melvin JA, McCafferty DG. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers 2010; 94(4):385-96. doi: 10.1002/bip.21472 [Crossref] [ Google Scholar]
- Paterson GK, Mitchell TJ. The biology of Gram-positive sortase enzymes. Trends Microbiol 2004; 12(2):89-95. doi: 10.1016/j.tim.2003.12.007 [Crossref] [ Google Scholar]
- Igarashi T, Asaga E, Goto N. The sortase of Streptococcus mutans mediates cell wall anchoring of a surface protein antigen. Oral Microbiol Immunol 2003; 18(4):266-9. doi: 10.1034/j.1399-302x.2003.00076.x [Crossref] [ Google Scholar]
- Semwal DK, Semwal RB, Combrinck S, Viljoen A. Myricetin: a dietary molecule with diverse biological activities. Nutrients 2016; 8(2):90. doi: 10.3390/nu8020090 [Crossref] [ Google Scholar]
- Chen W, Li Y, Li J, Han Q, Ye L, Li A. Myricetin affords protection against peroxynitrite-mediated DNA damage and hydroxyl radical formation. Food Chem Toxicol 2011; 49(9):2439-44. doi: 10.1016/j.fct.2011.06.066 [Crossref] [ Google Scholar]
- Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT. DOCK 6: impact of new features and current docking performance. J Comput Chem 2015; 36(15):1132-56. doi: 10.1002/jcc.23905 [Crossref] [ Google Scholar]
- Luo H, Liang DF, Bao MY, Sun R, Li YY, Li JZ. In silico identification of potential inhibitors targeting Streptococcus mutans sortase A. Int J Oral Sci 2017; 9(1):53-62. doi: 10.1038/ijos.2016.58 [Crossref] [ Google Scholar]
- Sivaramakrishnan M, Sharavanan VJ, Durairaj DR, Kandaswamy K, Piramanayagam S, Kothandan R. Screening of curcumin analogues targeting sortase A enzyme of Enterococcus faecalis: a molecular dynamics approach. J Proteins Proteom 2019; 10(3):245-55. doi: 10.1007/s42485-019-00020-y [Crossref] [ Google Scholar]
- Hejazinia F, Fozouni L, Azami NS, Mousavi S. The anti-biofilm activity of oregano essential oil against dental plaque-forming Streptococcus mutans in vitro and in vivo. J Kermanshah Univ Med Sci 2020; 24(3):e107680. doi: 10.5812/jkums.107680 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA: CLSI; 2020.
- Sarkar G, Turner RT, Bolander ME. Restriction-site PCR: a direct method of unknown sequence retrieval adjacent to a known locus by using universal primers. PCR Methods Appl 1993; 2(4):318-22. doi: 10.1101/gr.2.4.318 [Crossref] [ Google Scholar]
- Homonylo-McGavin MK, Lee SF. Role of the C terminus in antigen P1 surface localization in Streptococcus mutans and two related cocci. J Bacteriol 1996; 178(3):801-7. doi: 10.1128/jb.178.3.801-807.1996 [Crossref] [ Google Scholar]
- Sug-Joon Ahn, Sang-Joon Ahn, Zezhang T. Wen, L. Jeannine Brady, Robert A. Burne. Characteristics of Biofilm Formation by Streptococcus mutans in the Presence of Saliv. Infect Immun 2008; 76(9):4259-68. doi: 10.1128/IAI.00422-08 [Crossref] [ Google Scholar]
- Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 2016; 14(9):563-75. doi: 10.1038/nrmicro.2016.94 [Crossref] [ Google Scholar]
- Hejazinia F, Fozouni L. The anti-biofilm role of triclosan on the activity of streptococcal isolates containing sortase A in control of dental caries. J Neyshabur Univ Med Sci 2021;9(3):97-106. [Persian].
- Ce´line M, Le´vesque Le´vesque, Elena Voronejskaia, Yi-Chen Cathy Huang, Richard W. Involvement of Sortase Anchoring of Cell Wall Proteins in Biofilm Formation by Streptococcus mutans. Infect Immun 2005; 73(6):3773-77. doi: 10.1128/IAI.73.6.3773-3777.2005 [Crossref] [ Google Scholar]
- Sharavanan VJ, Sivaramakrishnan M, Kothandan R, Muthusamy S, Kandaswamy K. Molecular docking studies of phytochemicals from Leucas aspera targeting Escherichia coli and Bacillus subtilis subcellular proteins. Pharmacogn J 2019; 11(2):278-85. doi: 10.5530/pj.2019.11.43 [Crossref] [ Google Scholar]
- Doss CG, Chakraborty C, Chen L, Zhu H. Integrating in silico prediction methods, molecular docking, and molecular dynamics simulation to predict the impact of ALK missense mutations in structural perspective. Biomed Res Int 2014; 2014:895831. doi: 10.1155/2014/895831 [Crossref] [ Google Scholar]
- Zhulenkovs D, Rudevica Z, Jaudzems K, Turks M, Leonchiks A. Discovery and structure-activity relationship studies of irreversible benzisothiazolinone-based inhibitors against Staphylococcus aureus sortase A transpeptidase. Bioorg Med Chem 2014; 22(21):5988-6003. doi: 10.1016/j.bmc.2014.09.011 [Crossref] [ Google Scholar]
- Wallock-Richards DJ, Marles-Wright J, Clarke DJ, Maitra A, Dodds M, Hanley B. Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone. Chem Commun (Camb) 2015; 51(52):10483-5. doi: 10.1039/c5cc01816a [Crossref] [ Google Scholar]
- Huang P, Hu P, Zhou SY, Li Q, Chen WM. Morin inhibits sortase A and subsequent biofilm formation in Streptococcus mutans. Curr Microbiol 2014; 68(1):47-52. doi: 10.1007/s00284-013-0439-x [Crossref] [ Google Scholar]
- Yang WY, Won TH, Ahn CH, Lee SH, Yang HC, Shin J. Streptococcus mutans sortase A inhibitory metabolites from the flowers of Sophora japonica. Bioorg Med Chem Lett 2015; 25(7):1394-7. doi: 10.1016/j.bmcl.2015.02.051 [Crossref] [ Google Scholar]
- Hendrickx AP, Willems RJ, Bonten MJ, van Schaik W. LPxTG surface proteins of enterococci. Trends Microbiol 2009; 17(9):423-30. doi: 10.1016/j.tim.2009.06.004 [Crossref] [ Google Scholar]
- Cascioferro S, Raffa D, Maggio B, Raimondi MV, Schillaci D, Daidone G. Sortase A inhibitors: recent advances and future perspectives. J Med Chem 2015; 58(23):9108-23. doi: 10.1021/acs.jmedchem.5b00779 [Crossref] [ Google Scholar]
- Cascioferro S, Totsika M, Schillaci D. Sortase A: an ideal target for anti-virulence drug development. Microb Pathog 2014; 77:105-12. doi: 10.1016/j.micpath.2014.10.007 [Crossref] [ Google Scholar]
- Hu P, Huang P, Chen WM. Curcumin inhibits the Sortase A activity of the Streptococcus mutans UA159. Appl Biochem Biotechnol 2013; 171(2):396-02. doi: 10.1007/s12010-013-0378-9 [Crossref] [ Google Scholar]
- Kumar KM, Anbarasu A, Ramaiah S. Molecular docking and molecular dynamics studies on up beta-lactamases and penicillin binding proteins. Mol Biosyst 2014; 10(4):891-00. doi: 10.1039/c3mb70537d [Crossref] [ Google Scholar]