Avicenna Journal of Medical Biochemistry. 12(1):62-63.
doi: 10.34172/ajmb.2489
Letter to Editor
A New Linear Derivative of the Michaelis-Menten Equation
Mohammad Ali Zarei 1, * 
Author information:
1Department of Biological Sciences, Faculty of Science, University of Kurdistan, Sanandaj, Iran
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Zarei MA. A new linear derivative of the michaelis-menten equation. Avicenna J Med Biochem. 2024; 12(1):62-63. doi:10.34172/ajmb.2489
Dear Editor,
Years of experience in teaching biochemistry and enzymology courses, especially the discussion of enzyme kinetics, has enabled us to determine the kinetic parameters of enzymes, especially the Michaelis constant Km and the maximum reaction speed Vmax using various mathematical equations. These equations include the Michaelis-Menten equation and many of its linear derivatives such as Lineweaver-Burk, Woolf-Augustinsson-Hofstee (Eadie- Hofstee in some text books), Eadie-Scatchard, and Hanes-Woolf equations (1,2). However, it seems that none of the above linear graphs can directly and quickly calculate the kinetic parameters of the Michaelis constant Km and the maximum reaction speed Vmax from the line equation. This inspired us to think about a new linear derivative of the Michaelis-Menten equation, by which we can directly calculate the Michaelis constant Km and the maximum reaction rate Vmax from the equation of the graph line.
Considering the graphs of Woolf-Augustinsson-Hofstee (V0 vs. V0/[S0]), Eadie-Scatchard (V0/[S0] vs. V0), and Hanes-Woolf ([S0]/V0 vs. [S]), the need for a new plot can be felt, which shows the change in [S0] vs. [S0]/V0. Therefore, a diagram was drawn and its equation was extracted. Accordingly, the path of its derivation from the Michaelis-Menten equation is as follows:
-
V0 = (Vmax [S0])/Km + [S0],
-
V0Km + V0[S0] = Vmax[S0],
-
(V0Km)/V0 + (V0[S0])/V0 = Vmax[S0]/V0,
-
Km + [S0] = Vmax[S0]/V0,
-
[S0] = Vmax[S0]/V0 - Km
In equation 5, which is related to the linear graph of changes of [S0] against [S0]/V0, the value of the maximum reaction speed Vmax is directly obtained from the slope of the graph and the line constant by changing the sign of Michaelis constant Km. As an example, we considered a problem presented by Mikkelsen and Cortón (3).
The information in the first and second rows of Table 1 was obtained from an enzyme saturation experiment by the substrate, determine the values of Michaelis constant Km and the maximum reaction rate Vmax by drawing a diagram.
Table 1.
Substrate Concentration Values and Enzyme Reaction Rates Measured for Them
|
1
|
[S0] mM
|
0.368 |
0.448 |
0.568 |
0.800 |
1.336 |
4 |
|
2
|
V0 μmol/min
|
0.275 |
0.310 |
0.353 |
0.418 |
0.488 |
0.648 |
|
3
|
1/[S0]
|
2.717 |
2.232 |
1.760 |
1.250 |
0.748 |
0.250 |
|
4
|
1/V0
|
3.636 |
3.226 |
2.832 |
2.392 |
2.049 |
1.5432 |
|
5
|
[S0]
|
0.368 |
0.448 |
0.568 |
0.800 |
1.336 |
4 |
|
6
|
[S0]/V0
|
1.338 |
1.445 |
1.609 |
1.913 |
2.737 |
6.172 |
At first we convert the raw data to the form which can be used in Lineweaver-Burk (rows 3 and 4 of Table 1) and new method (rows 5 and 6 of Table 1).
After drawing the corresponding graphs (Figures 1a and 1b), the values of Michaelis constant Km and the maximum reaction rate Vmax can be calculated from the equation of the line of each graph.
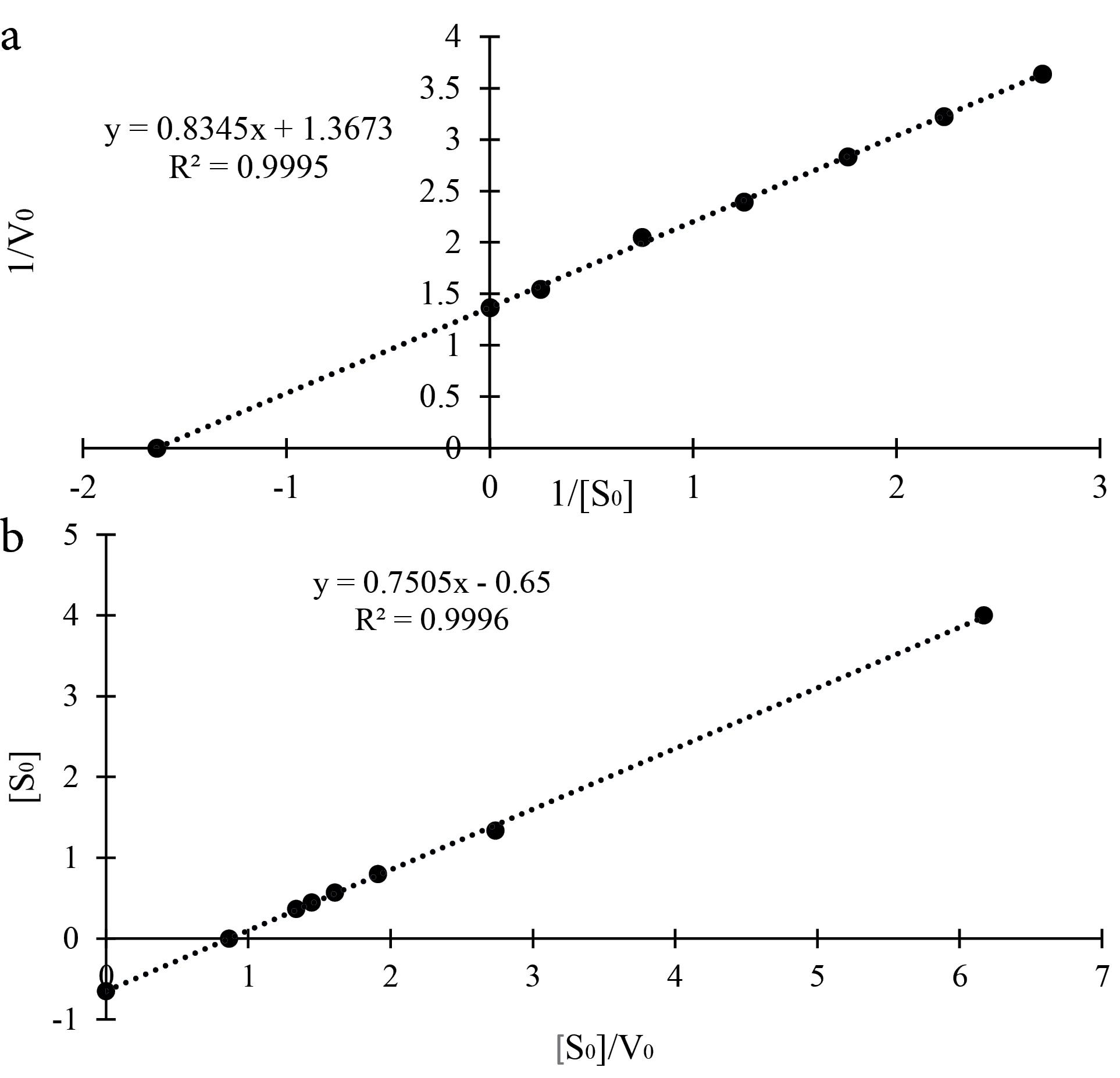
Figure 1.
a) Lineweaver-Burk Plot of Changes of 1/V0 vs. 1/[S0] b) New Plot, Changes of [S0] vs. [S0]/V0
.
a) Lineweaver-Burk Plot of Changes of 1/V0 vs. 1/[S0] b) New Plot, Changes of [S0] vs. [S0]/V0
The line equation of the Lineweaver-Burk diagram is as follows:
1/V0 = (Km/Vmax).1/[S0] + 1/Vmax
y = 0.8345x + 1.3673
As a result, 1/Vmax = 1.3673, so Vmax = 0.73 μmol/min.
On the other hand, Km / Vmax = 0.8345, as a result 0.73136 / Km = 0.8345 and then Km = 0.61 mM.
The equation of the line of the new graph is as follows:
[S0] = Vmax[S0]/V0 - Km
y = 0.7505 x – 0.65
As a result, Vmax = 0.75 μmol/min.
On the other hand, -Km = -0.65, as a result, Km = 0.65 mM.
The slight difference that was observed in the Michaelis constant Km and the maximum reaction speed Vmax obtained from the two approaches in the above working example is apparently caused by the unequal distance of the points due to the selection of certain concentrations of the substrate that are more suitable for the Lineweaver-Burk method. However, at non-selective concentrations of the substrate suitable for the Lineweaver-Burk plot, we can get more accurate results for the new method.
The new diagram can quickly give us the values of the Michaelis constant Km and the maximum reaction speed Vmax. The relation between this plot and the Hanes-Woolf plot is just similar to the relation between the Woolf-Augustinsson-Hofstee plot and the Eadie-Scatchard plot (1), in that we can go from one plot in the pair to the other just by changing the labels of y and x axes with each other. Therefore, the necessity of the proposed plot is equal to that of Woolf-Augustinsson-Hofstee plot. On the other hand, this plot is similar to the other three above-mentioned plots. However, in comparison with a double reciprocal plot like the Lineweaver-Burk plot, it is more straightforward in data analysis and result presentation.
A new single-reciprocal plot was derived from the Michaelis-Menten equation, which can be used for enzymatic analysis along with previous plots, and it is possible to directly calculate the Km and Vmax values from the equation of the plot without any additional mathematical operation.
Competing Interests
There is no competing interest.
Funding
None.
References
- Segel IH. Biochemical Calculations. 2nd ed. John Wiley & Sons; 1976. p. 205-323.
- Palmer T, Bonner PL. Enzymes: Biochemistry, Biotechnology, Clinical Chemistry. 2nd ed. Woodhead Publishing; 2007. p. 105-25.
- Mikkelsen SR, Cortón E. Bioanalytical Chemistry. 1st ed. John Wiley & Sons; 2016. p. 83-60.