Avicenna Journal of Medical Biochemistry. 12(1):30-38.
doi: 10.34172/ajmb.2524
Review Article
Proteomic Alterations and Metabolic Pathway Modulations Induced by Methyl Methane Sulfonate Treatment in Response to DNA Damage
Mohd Mustafa 1  , Badar ul Islam 2, Irfan Qadir Tantry 3, Moinuddin 1, Safia Habib 1, *
, Badar ul Islam 2, Irfan Qadir Tantry 3, Moinuddin 1, Safia Habib 1, * 
Author information:
1Department of Biochemistry, Jawaharlal Nehru Medical College, Faculty of Medicine, Aligarh Muslim University, Aligarh, India
2Department of Biochemistry, Ram Gopal Medical College and Research Center, Hathras, India
3Department of Biochemistry, School of Biological Sciences, University of Kashmir, Srinagar, India
Abstract
Exposure to methyl methane sulfonate (MMS), an alkylating agent, induces DNA damage, increasing cellular and metabolic sensitivity, leading to cellular demise and a delay in the cell cycle. This delay is attributed to changes in global transcription regulation, resulting in significant alterations in the proteome. Numerous studies have focused on transcriptome changes post-MMS treatment. However, cellular proteomic changes remain underexplored. This brief literature review is based on the assessment of studies obtained from several PubMed, Web of Science, and Scopus databases and Google Scholar using specific keywords. The article is based on manuscripts published between 2005 and 2024. Proteomic analysis identified 53 proteins, with 36 upregulated, 10 downregulated statuses, and a few exhibiting no or negligible changes. MMS exposure reflected changes in the phosphorylation status of the carboxy-terminal domain (CTD) of RNA polymerase II (RNAP-II) and a gross change in the transcriptome. Literature also supports the proteome change of the RNAP-II complex and ~1640 peptides, corresponding to 27 interacting proteins and the twelve RNAP-II subunits. The identified proteins were involved in DNA repair and energy pathway modulation. Notably, significant changes were observed in enzymes mostly related to carbohydrate and amino acid metabolism, predominantly, glycolysis, the fate of pyruvate, and the biosynthetic pathways of amino acid metabolism. The available literature supports a co-regulated response to MMS-induced DNA damage involving both DNA repair mechanisms and metabolic pathways.
Keywords: Methyl methane sulfonate, DNA damage, Proteome changes, Metabolic pathways, Transcription regulation, Oxidative stress,
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Mustafa M, Islam BU, Tantry IQ, Moinuddin, Habib S. Proteomic alterations and metabolic pathway modulations induced by methyl methane sulfonate treatment in response to DNA damage. Avicenna J Med Biochem. 2024; 12(1):30-38. doi:10.34172/ajmb.2524
Background
Methyl methane sulfonate (MMS) is a mutagenic chemical. It is a commonly used laboratory compound, an industrial solvent, and an environmental toxicant with a well-established function for DNA alkylation. The alkylation of DNA in this case leads to N-alkylated base products. The response of an MMS attack on double-stranded DNA is to produce 7-methylguanine (m7G, about 80%) and 3-methyladenine (m3A, about 10%). However, interaction with single-stranded DNA leads to the accumulation of 10% 1-methyl adenine and 8% 3-methyl cytosine (1). In addition to being an alkylating agent, MMS is a known carcinogen (2). It methylates oxygen and nitrogen atoms of the nitrogen-containing bases at the N3 and N7 positions of deoxy-adenosine and deoxy-guanosine, respectively, and perturbs the DNA backbone (3), resulting in single-strand breaks, which could then lead to double-strand breaks during the S-phase of the cell cycle. Exposure to these genotoxic agents induces DNA damage, stimulating cellular responses to perpetuate genomic integrity. Cells must maintain the stability and coherence of their genomes to consistently pass on genetic information from one generation to the next. MMS-induced alkylation modifies DNA bases, resulting in mutations and potentially triggering cellular apoptosis or necrosis (4). A key consequence of MMS-induced DNA damage is a delay in the cell cycle, primarily due to alterations in comprehensive global transcription regulation and subsequent changes and alterations in the cellular proteome pool (5-7). The mentioned changes are potential sources of genome instability that can induce cell death, tumor development, malignant transformations, and other biological complications as well. If these DNA lesions escape the repair mechanism or are handled by the pathways that could not correct MMS-induced structural or conformational perturbations, it could result in the accumulation of modified DNA products, qualitatively or quantitatively altered translation products, and immune complexes (i.e., a likely cause of carcinogenesis). Understanding cellular response toward MMS may contribute to enhancing the knowledge of DNA methylations, the cellular repair mechanisms, and the impact of this environmental and laboratory toxin on the induction of chronic ailments such as metabolic syndrome, diabetes mellitus, cancers, and other acquired health issues. This issue paves the way for future investigations and potential therapeutic interventions. An outline of the effects of MMS exposure in the cellular system is depicted in Figure 1.
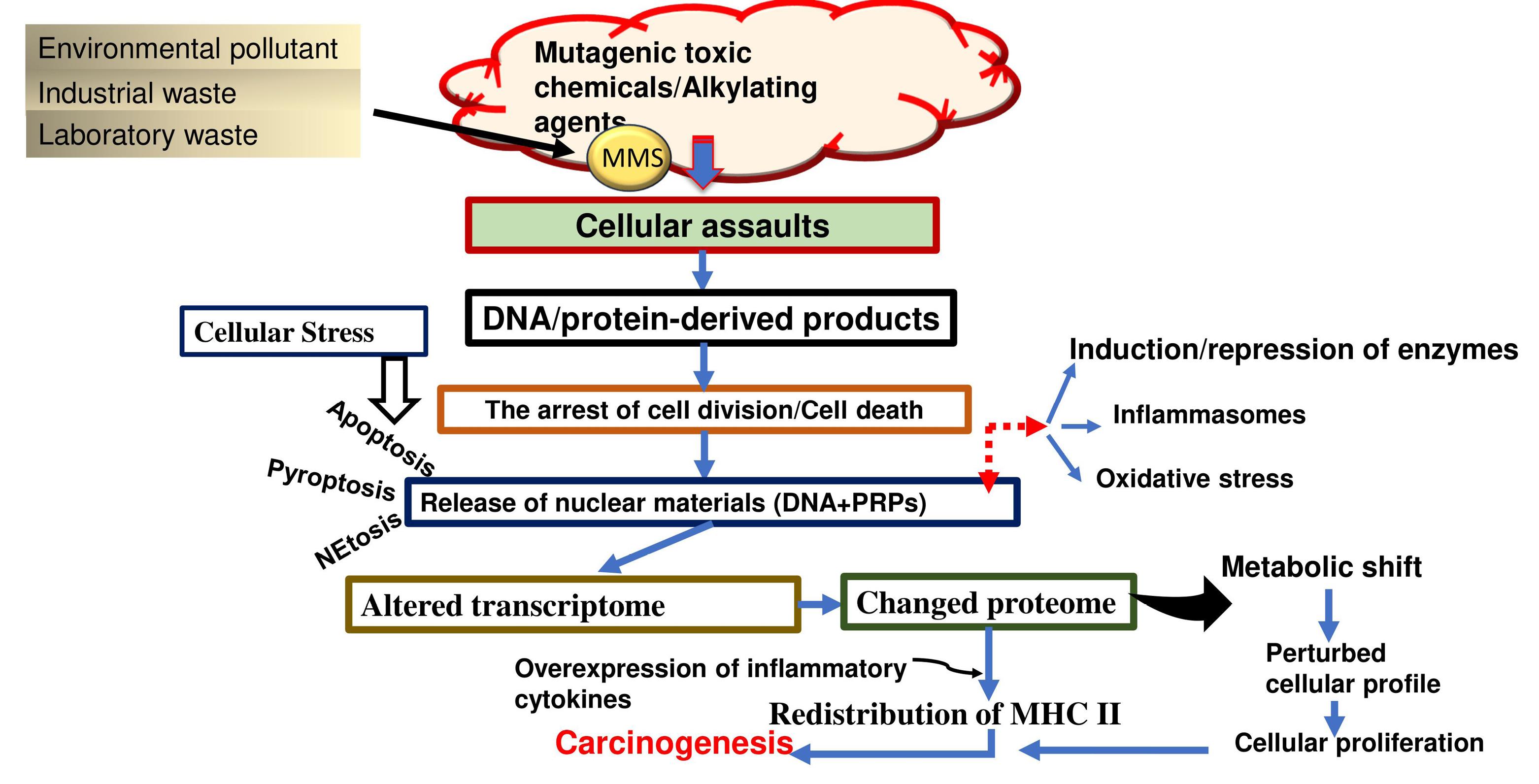
Figure 1.
The figure represents an overview of cellular responses upon MMS exposure. The MMS assaults lead to a delayed cell cycle, metabolic shift, oxidative stress, and a pro-inflammatory response culminating in chronic conditions and cancers.
.
The figure represents an overview of cellular responses upon MMS exposure. The MMS assaults lead to a delayed cell cycle, metabolic shift, oxidative stress, and a pro-inflammatory response culminating in chronic conditions and cancers.
As discussed above, MMS is known to cause cytotoxic and genotoxic effects, mainly by disrupting the DNA at the structural and conformational levels. It is also reported to induce oxidative stress (OS) in the cellular system. This article aims to explore the proteomic changes that occur inside the cell exposed to MMS and to specifically identify the effect of MMS exposure on the metabolic profile of the MMS-exposed system for a better understanding of metabolic tolerance.
A Brief Overview and Action Mechanism of Methyl Methane Sulfonate
Most of the methylating agents target DNA. This property is mediated by electrophilicity and acts either through SN1 or SN2 type of reactions. Here, MMS is the typical SN2 types of compound, exhibiting high affinity toward the nitrogen atoms of purines and pyrimidines. This attack on DNA results in adduct formation in the double-stranded DNA (8,9). Out of many nucleobases, N7-methylguanine has a high concentration. Although this adduct does not interfere with DNA replication, it is still cytotoxic and genotoxic due to the production and accumulation of apurinic sites formed by depurination.
Along with N7-methyl guanine, other minor adducts that are formed also lead to increased production of apurinic sites, altered transcriptomes, and proteomes (4,6). The minor adducts potentially include N3-methyladenine, N3-methylguanine, and, to the same extent, O6-methylguanine. These minor adducts are pretty unstable and undergo hydrolysis. MMS follows an intermediate transition state since it represents a classical SN2 type of compound model. Therefore, it has a high Swain-Scott constant (S-value > 0.83) targeting nucleophilic centers, such as N3 adenine and N7 guanine. However, MMS is mentioned to be a relatively weak mutagen because the major adduct formed, N7-methyl guanine, is not mutagenic but becomes mutagenic after forming a ring-opened adduct. This creates primary sites handled by base excision repair, contributing to the genotoxic threshold of MMS.
Both of these methylated bases are known to induce cytotoxicity, interfere with the replication fork, and cause G→A transitions. As far as O6-methyl-2-deoxyguanine is concerned, it is usually tackled by O6-methylguanine DNA methyl transferases (MGMT). N3-methyl-2-deoxyadenosine, being unstable, quickly depurinates to produce abasic sites (AP). The lesions induced by N3-methyl-2-deoxyadenosine are reported to block both α and γ DNA polymerases, whereas DNA polymerases η and κ are reported to bypass the AP lesions (10). N3-methyl-2-deoxyadenosine has a half-life of 12‒24 hours and, as mentioned earlier, is quite unstable; therefore, its biochemical and biophysical quantifications appear to be a not-so-good target (11). Based on this information, it is expected that MMS exposure may result in metabolic shifts due to DNA damage-induced proteomic disruption.
Materials and Methods
The literature review is based on the search performed through PubMed, Web of Science, and Scopus databases and Google Scholar using keywords such as “methyl methane sulfonate”, “Methyl methane sulfonate + DNA damage”, “Methyl methane sulfonate + cellular proteome”, “methyl methane sulfonate + cancer”, “methyl methane sulfonate + oxidative stress”. The article is based on manuscripts published between 2005 and 2024 in the English language. All the duplicate articles were removed, and finally, 44 manuscripts were selected for this brief review. English articles specifically discussing MMS toxicity and cellular proteomic alterations impacting the cellular metabolic profile were included in the study. However, articles in languages other than English, articles published before 2005, duplicate articles, and those not focusing on MMS-induced cytotoxicity, genotoxicity, and metabolic protein profile were excluded from the study.
Cell Cycle Delay and Global Transcription Regulation
Due to the MMS-induced assaults on the DNA, aberrant DNA damage products accumulate, and an orchestrated cell cycle progression critical for cell duplication is at fault. The stress-induced attenuation of DNA replication, mainly due to exogenous chemical exposures, causes cell cycle arrest at checkpoints, which, in turn, compromises repair mechanisms. G1-S transition points typically become non-functional in malignant cells, specifically in the case of breast and ovarian cancers. To compensate and maintain genetic integrity, distinct proteins involved in cell cycle checkpoint regulation are translated, and the activation of DNA repair pathways is initiated (5). Upon MMS treatment, cells exhibit a pronounced delay in the cell cycle, triggered by DNA damage. This delay appears to be an acquired protective mechanism that allows the exposed cells to have a buffer time to repair the inflicted molecular nucleic acid damage (5-7). To speed up the repair process, the cell cycle is forced to undergo several modulations at checkpoints and rectify different points of control, including the DNA damage checkpoint, transcriptional modifications, replication fork stalling, and, at times, cell cycle arrest. The downstream effector kinase Rad53 and adaptors Rad9 or Mrc1 participate in the traditional checkpoint activation pathway mediated by the sensor kinase Mec1, which phosphorylates upon activation. Additionally, the transcription of DNA damage response (DDR) genes, specifically NBS1, ATM, BRCA1, BRCA2, FANCD2, and dNTP pools, is regulated by active Rad53, either directly or through Dun1 dependence (12,13). The delay is orchestrated by changes in transcription regulation. The overall effect is presented by a significant alteration in the proteome level. These transcriptional changes are detected at the transcriptome level through various microarray studies, but the detection and identification of proteomic changes are arduous tasks and have been less extensively studied so far.
RNA Polymerase II and Protein Interactions
MMS-induced DNA methylations result in an alteration in the phosphorylation status of the carboxy-terminal domain (CTD) of RNA polymerase II (RNAP-II). The RNAP-II complex facilitates the transcription of mRNA in all eukaryotes. The CTD is made up of repeated units of Y1S2P3T4S5P6S7 consensus sequences and is found in the largest subunit of RNAP-II (Rpb1). Numerous posttranslational changes to the CTD cause a wide range of proteins to associate and dissociate with it, which is necessary for active transcription (14,15). In response to DNA damage, RNAP-II phosphorylates more CTDs, particularly Ser2, and factors mediating or responding to this phosphorylation regulate survival in DNA damage-affected environments (16). The CTD becomes hyperphosphorylated as a result of alterations in Ser2, 5, and 7. The transcription initiation, elongation, termination, mRNA splicing, and mRNA export are all regulated by the phosphorylation and dephosphorylation of CTD. It also controls the binding of other transcriptional regulatory proteins to RNAP-II (16,17). Therefore, RNAP-II plays a crucial role in transcription, and changes in its phosphorylation status can impact transcriptional regulation. Transcription regulatory proteins are recruited with the aid of the CTD. A study reported and identified 1640 peptides corresponding to 27 interacting proteins, along with the involvement of twelve subunits of RNAP-II through an affinity purification of the RNAP-II complex, followed by nano-LC/MS analysis (6).
Proteome Changes Post-Methyl Methane Sulfonate Treatment
A comprehensive proteomic analysis revealed changes in protein expression and interaction networks following MMS treatment. The analysis by Bharati et al (6) identified 53 proteins using mass spectrometry (MS/MS), of which 36 were upregulated, 10 were downregulated, and a few exhibited insignificant changes (Table 1). They also reported that MMS-induced changes in the protein profile mostly belong to the basal transcription factors that include Tgf1/2, Spt5, Irw1, and Rtr1, while Rvb1/2, Nop1/58, and Cbf5 belong to RNA biogenesis. The basal transcription factors and RNA biogenesis factors are responsible for generalized global transcription regulation. A study by Kim et al (18) identified DDR-associated proteins, including Vma1/2/4, Stm1, Cys3, and Tif1, to be expressed under MMS stress. The study also reported that the level of Rnr4, Vma1/2, Stm1, and Pre9 proteins was highly expressed, while Vma4, Bmh1, Cys3, and Tif1 showed a slight change in protein expression.
Table 1.
List of Proteins Identified to Undergo Methylation-Induced Proteomic Changes
|
Pathway Involved
|
Genes Affected
|
Proteins Modulated
|
Type of Regulationa
|
Times Fold Changeb
|
| Glycolysis (19,20) |
Fba1
|
Fructose 1,6-bisphosphate aldolase |
Up-regulation |
1.46-2.47 |
|
Pgi1
|
Phospho-gluco-isomerase |
Up-regulation |
1.53 |
|
Hxk2
|
Hexokinase isoenzyme 2 |
Up-regulation |
2.23 |
|
Tdh2
|
Triose-phosphate de hydrogenase |
Up-regulation |
2.09-2.47 |
|
Tdh3
|
Triose-phosphate de hydrogenase |
Up-regulation |
3.20 |
|
Gpm1
|
Tetrameric phosphoglycerate mutase |
Up-regulation |
1.22 |
|
Eno1
|
Phosphopyruvate hydratase |
Up-regulation |
3.27 |
|
Eno2
|
Phosphopyruvate hydratase |
Up-regulation |
1.97-3.27 |
|
Pgk1
|
3-PhosphoGlycerate kinase |
Up-regulation |
1.68 |
|
Tpi1
|
Triose phosphate isomerase |
Down-regulation |
0.86 |
|
Pdc1
|
Pyruvate decarboxylase |
Up-regulation |
2.67 |
|
Adh1
|
Alcohol dehydrogenase |
Up-regulation |
2.56-3.54 |
| Pentose phosphate pathway (6,21) |
Tkl1
|
Transketolase |
Up-regulation |
2.03-3.97 |
|
Gnd1
|
6-phosphogluconate dehydrogenase |
Up-regulation |
1.80 |
| Amino acid and nucleotide metabolism (6,22) |
Gdh1
|
Glutamate dehydrogenase |
Down-regulation |
0.72 |
|
Ilv5
|
Acetohydroxyacid reductoisomerase |
Up-regulation |
1.83 |
|
Cys3
|
Cystathionine gamma-lyase |
Up-regulation |
1.2 |
|
Cys4
|
Cystathionine beta-synthase |
Up-regulation |
2.47 |
| Protein biosynthesis (6,18) |
Frs1
|
Phenylalanyl (F)-tRNA synthetase |
Down-regulation |
0.74 |
|
Tif1
|
Translation initiation factor eIF4A |
Up-regulation |
3.52 |
|
Rpp0
|
Ribosomal Protein P0 |
Up-regulation |
9.17 |
|
RPS0A
|
Ribosomal 40S subunit protein S0A |
Down-regulation |
0.88 |
| Cell cycle and growth regulation (6) |
Bmh1
|
Brain Modulo signaling homologue |
Up-regulation |
2.92 |
|
Stm1
|
Suppressor of ToM1 |
Up-regulation |
2.26 |
|
Vma1
|
Vacuolar membrane ATPase |
Up-regulation |
3.30 |
|
Vma2
|
Vacuolar membrane ATPase |
Up-regulation |
2.88 |
|
Rnr4
|
Ribonucleotide reductase |
Up-regulation |
8.41 |
|
Vma4
|
Vacuolar H + -ATPase |
Down-regulation |
0.75 |
a Fold change in gene expression is reported to be dependent on MMS concentration.
b Source. Bharati et al (6).
Proteins were mostly associated with basal transcription factors, and changes were reported to correspond to Saccharomyces cerevisiae after concentration-dependent MMS treatment. Another analysis demonstrated 306 distinct genes whose transcription was substantially impacted by MMS (7). The identified proteins were categorized into several functional groups, including cell cycle modulators, metabolic enzymatic components, and macromolecules involved in protein folding and organization (18).
Impact on Metabolic Pathways
The proteomic analysis revealed substantial changes in the key metabolic enzymes and regulators of glycolysis, pyruvate metabolism, and amino acid biosynthesis. These enzymes are integral to central cellular energy production and the synthesis of paramount biomolecules and their derivatives. The observed changes suggest that DNA damage induced by MMS not only affects DNA repair mechanisms but also has a broader impact on the cellular metabolic milieu. This co-regulation indicates a tightly linked relationship between DDR and metabolic pathway regulation (Figure 2). Malignant cells possess the signature characteristic of rapid proliferation, which requires alterations to tumor metabolism and its microenvironment. Metabolic shifts must be enough to meet the elevated bioenergetic demands. This forms the basis of an upcoming concept termed ‘deregulating cellular energetics’ and stands as a new hallmark of cancer metabolism. Cancer cells present with a distinct metabolic phenotype compared to noncancerous cells. These acquired changes in metabolic profiles and fluxes are achieved by shifts in the cellular transcriptome and proteome, as well as extensive changes in metabolic reprogramming for uncontrolled anabolic growth. Broadly, this involves overcoming the nutritional scarcity and maintaining cellular uptake of simple nutrients (glucose, amino acids, and the like), which are channeled into energy-extracting pathways and biosynthetic metabolic sequences via the core major metabolic pathways of glycolysis, the Krebs cycle, the hexose monophosphate shunt, and the pathways directed for the synthesis of non-essential amino acids. Finally, converging to the ATP-dependent complex formation of biological macromolecules is ultimately required for the rapid proliferation of a neoplastic cell (23,24). The outcome demonstrated the interactive relationship between phosphatase (Rtr1) and kinase (Ire1) with RNAP-II. Rtr1 is a phosphatase that aids in the dephosphorylation of the CTD’s Ser5 site, which aids in the elongation and termination of transcription. On the other hand, Irw1 was found to be a component of the complex that moved the RNAP-II complex to the nucleus, where transcription factors are involved (25,26). The recruitment of these proteins may activate global transcription, which in turn may control the range of genes involved in the MMS-induced OS.
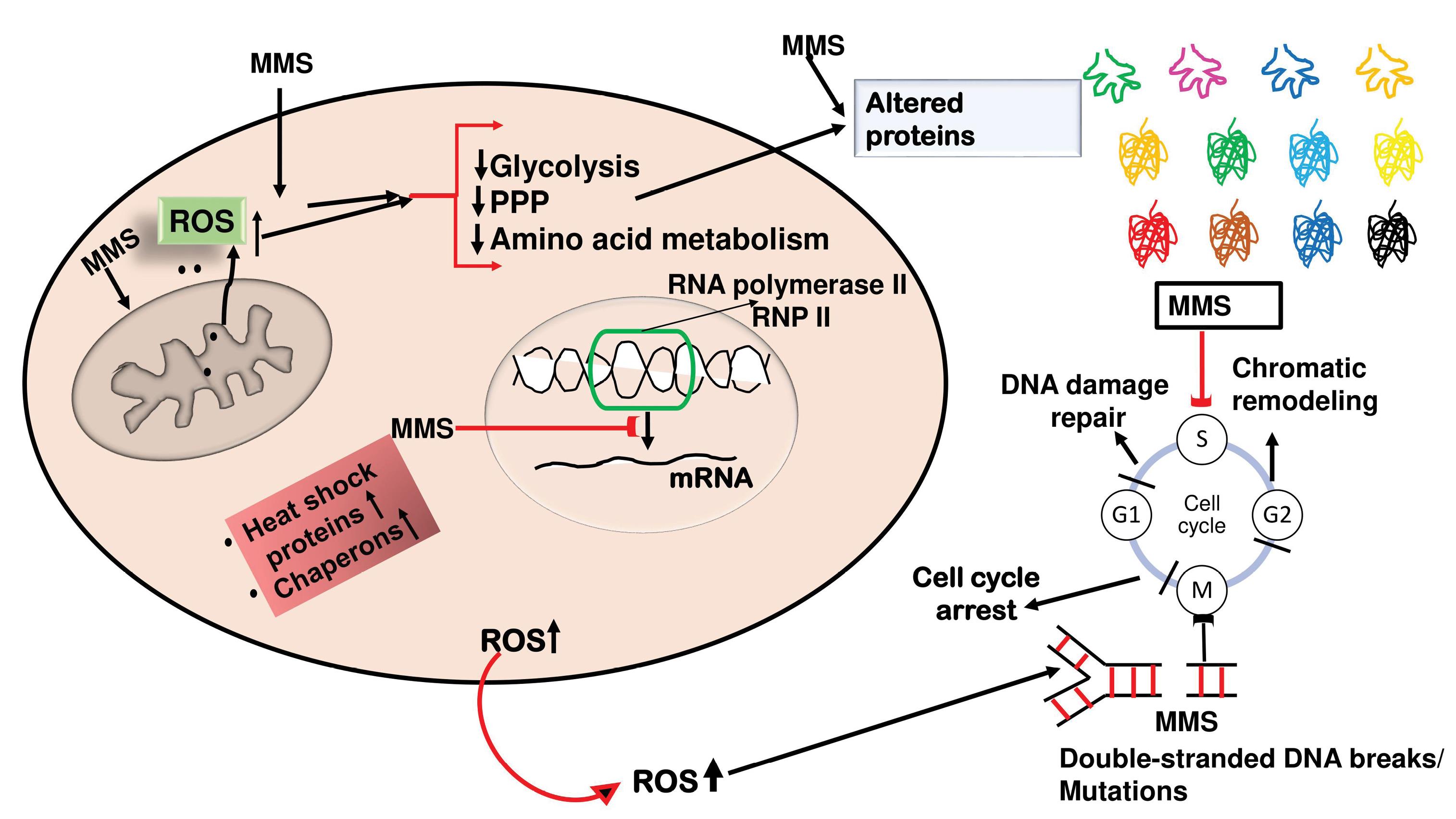
Figure 2.
A simplified schematic representation of methyl methane sulfonate (MMS) induced cellular damage and generation of reactive oxygen species (ROS), which leads to a decline in metabolic pathways like glycolysis, pentose phosphate pathways (PPP), and amino-acid metabolism. These effects are reflected in altered/deranged proteomics at the cellular level. The ROS also leads to increased synthesis of chaperones and heat shock proteins to rectify the damage to folded proteins (leading to altered proteomics). The ROS and MMS also induce DNA damage, and double-stranded breaks, which prolong the DNA repair mechanisms, and chromatin remodeling finally leads to cell cycle arrest. MMS also binds to the c-terminal end of RNA pol II and inhibits mRNA transcription.
.
A simplified schematic representation of methyl methane sulfonate (MMS) induced cellular damage and generation of reactive oxygen species (ROS), which leads to a decline in metabolic pathways like glycolysis, pentose phosphate pathways (PPP), and amino-acid metabolism. These effects are reflected in altered/deranged proteomics at the cellular level. The ROS also leads to increased synthesis of chaperones and heat shock proteins to rectify the damage to folded proteins (leading to altered proteomics). The ROS and MMS also induce DNA damage, and double-stranded breaks, which prolong the DNA repair mechanisms, and chromatin remodeling finally leads to cell cycle arrest. MMS also binds to the c-terminal end of RNA pol II and inhibits mRNA transcription.
Altered Cellular Proteome Under Methyl Methane Sulfonate-Induced Oxidative Stress
The effect of MMS-induced OS on the cellular proteome is one of the important research aspects in molecular biology. This is because research on this topic sheds light on the molecular mechanisms that underlie DDR, repair mechanisms, and the overall cellular stress response (27). As mentioned, MMS is an alkylating chemical known to cause base mispairing, strand breaks, and cytotoxic lesions in DNA by forming methyl adducts, mainly at the N7-guanine and N3-adenine sites and replication stall sites. In response to such pronounced DNA damage, the cell adopts a complex SOS response that includes extensive modifications to cellular proteostasis and direct repair processes (4).
The activation of DDR pathways is one of the critical cellular responses to OS generated by MMS. Key proteins, including ataxia-telangiectasia mutated, ATM and Rad3-related, and DNA-dependent protein kinase catalytic subunit, are involved in orchestrating a complex network of signaling cascades of cellular response. These kinases phosphorylate downstream effectors, such as CHK1/2 and p53, which in turn regulate apoptosis in the event of irreversible damage, DNA repair, and cell cycle arrest. Thus, these extensive cellular defense systems are reflected in proteome changes under stress caused by MMS (28).
Mass spectrometry-based proteomic investigations have reported that OS caused by MMS induces notable alterations in the quantity and state of modification of a broad range of proteins. Heat shock proteins (HSPs) such as HSP70 and HSP90 are notably elevated. Assisting the correct folding of translated proteins, refolding the misfolded proteins, and tagging and designating irreparably damaged proteins for disintegration, these are the processes served by molecular chaperones for maintaining protein homeostasis. The increased cellular stress response intended to prevent the aggregation of damaged proteins is shown by the overexpression of HSPs (29).
A noteworthy reported modification has been related to the ubiquitin-proteasome system, which is in charge of specifically degrading proteins or protein products that are broken or misfolded. The expression and activity of certain proteins, including proteasomal subunits and ubiquitin ligases (E3 ligases), increase under MMS treatment. This increased proteolytic activity appears to be intended to remove damaged proteins and their misfolded by-products and forestall proteotoxic stress. Simultaneously, autophagy-related proteins such as LC3 and p62/SQSTM1 are reported to be upregulated, emphasizing the cell’s attempt to maintain proteostasis (30).
OS is known to have an impact on the antioxidant cellular defense system, causing changes in the gene expression levels of important enzymes such as glutathione peroxidase, catalase, and superoxide dismutase. These enzymes play a critical role in the neutralization of reactive oxygen species, cutting back OS and resulting oxidative damage by MMS. The attempt of the cell to counterbalance the increased oxidative burden and shield the vital cellular constituents, such as proteins, lipids, and nucleic acids, from oxidative damage is highlighted by the overexpression of these enzymes (31).
Another study corroborates the above findings reported by Bharati et al, demonstrating that OS caused by MMS profoundly modifies the metabolic landscape of the cell. Differential and dynamic expression of glycolytic enzymes, including pyruvate kinase, phosphofructokinase, and hexokinase, suggests a change in cellular metabolism (32). This metabolic reprogramming and altered proteome profile reflect the cell’s way of adjusting to respond to its enhanced energy needs to overcome stress and initiate and sustain repair processes. Furthermore, oxidative phosphorylation and tricarboxylic acid cycle enzymes exhibit altered expression and activity, indicating a more comprehensive metabolic adaptation to maintain appropriate cellular energy levels and prevent a state of deficit (33).
Additionally, modifications to the expression and structure of proteins implicated in DNA repair pathways have been found using proteomic analysis. DNA glycosylases, AP endonuclease, and DNA polymerase β are examples of base excision repair proteins that are increased to aid in the repair of methylated bases and strand breaks (34). Similarly, heightened expression of the components of the nucleotide excision repair pathway, including XPA, XPC, and ERCC1, signifies their function in repairing botched bulky protein adducts and organizational helix-distorting lesions caused by MMS (35).
Furthermore, there are reports of differential upregulations of proteins, including RAD51, BRCA1, and Ku70/Ku80, which are implicated in the non-homologous end joining and homologous recombination pathways (36). The effective repair of the double-strand break, one of the most detrimental DNA damages, is ensured and repaired by this control. The coordinated upregulation of various genes and their repair protein products demonstrates the cell’s diverse strategy for preserving genomic integrity for cellular sustenance during stress conditions induced by MMS (37,38).
MMS-induced OS also has a profound impact on post-translational modifications of proteins, including glycosylation, phosphorylation, ubiquitination, and acetylation (39). Numerous phosphorylation events on DDR proteins have been found by phosphoproteomic investigations, which alter the proteins’ native state, stability, and domain function. One such reported example is the phosphorylation of histone variant H2AX (γH2AX), which functions as a marker for double-strand breaks and aids in the recruitment of repair proteins to damaged sites (40). These proteins ensure appropriate coordinated DNA repair before cell division. Hence controlling the correct course of the cell cycle and monitoring ubiquitination and subsequent degradation of cyclin-dependent kinase inhibitors such as p21/27 (41).
Moreover, the expression of transcription factors is indicative of the cell’s proteome alteration under MMS-induced OS (42). A response to MMS stress is phosphorylation and acetylation to stabilize and activate the tumor suppressor protein p53, which triggers the transcription of genes linked to apoptosis, DNA repair, and cell cycle arrest. The interaction between DDR and inflammatory signaling is further highlighted by the activation of the nuclear factor-kappa B pathway, which controls the expression of genes involved in inflammation and immune response (43,44).
In conclusion, during MMS-induced OS, the cellular proteome experiences significant modification, which is indicative of the activation of a wide range of defense mechanisms meant to preserve cellular homeostasis, genomic integrity, and survival. These proteome modifications include changes in protein expression, modifications, and functions, impacting metabolic adaptation, DNA repair, protein quality control, and cell death pathways. Gaining knowledge of these proteome changes highlights the complex web of chemical reactions that protect cellular integrity and offers vital insights into the cellular defense mechanisms used to deal with oxidative damage caused by alkylation (Figure 3).
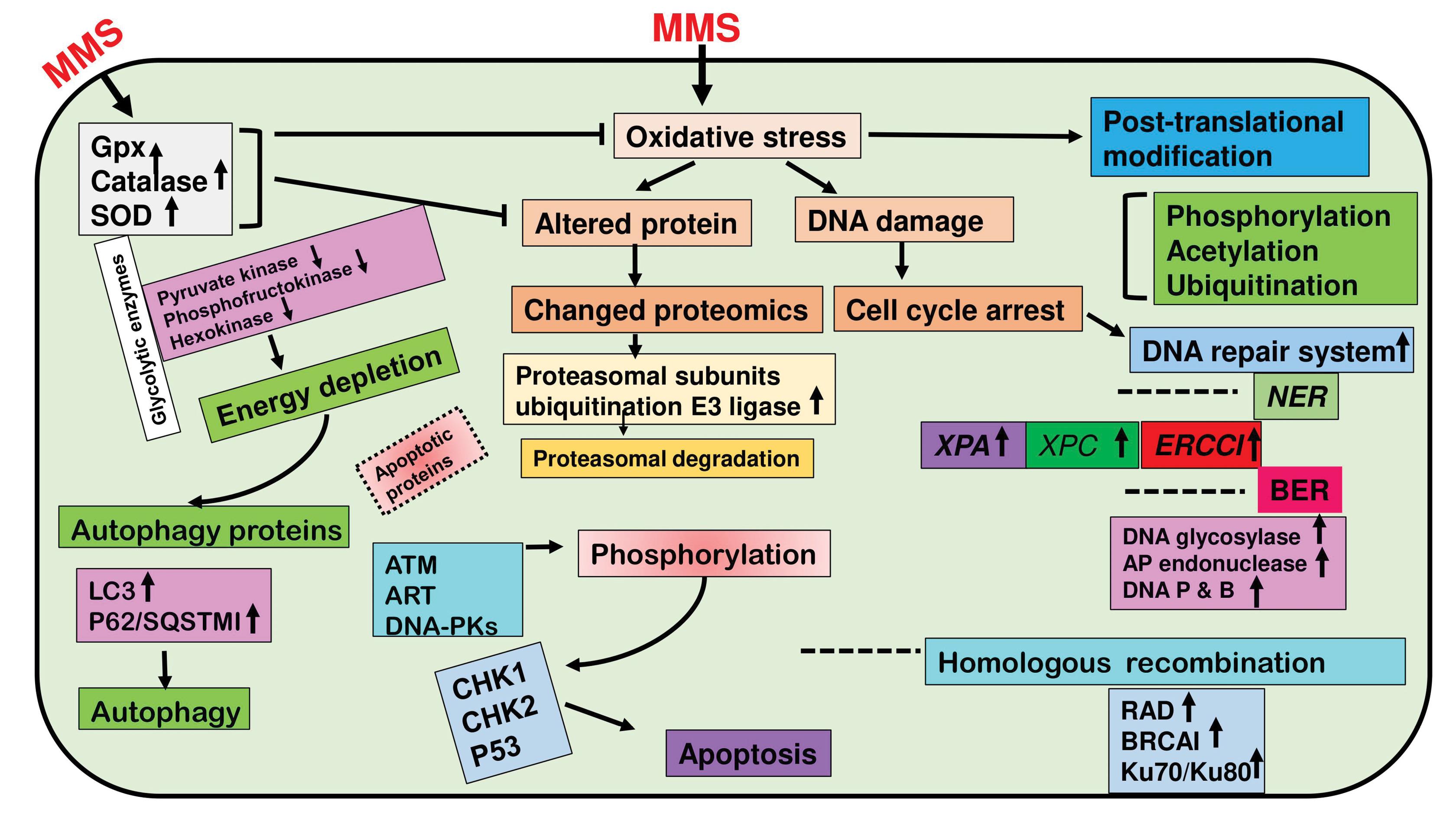
Figure 3.
Schematic diagram showing methyl methane sulfonate (MMS), increases reactive oxygen species (ROS) production which leads to oxidative stress that causes protein and DNA damage. The cell counters the stress response by increasing the expression of proteasomal degradation machinery. The cell also during the stress increases the activity of protein kinases (PKs) like ATM, ART, and DNA PKcs which activate CHK1 and CHK2 (DNA damage signaling proteins) and these then phosphorylate p53, which leads to apoptosis of the stressed cell. MMS-stressed cells also increase the expression of LC3 and P62/SQTM1 which leads to autophagy.
.
Schematic diagram showing methyl methane sulfonate (MMS), increases reactive oxygen species (ROS) production which leads to oxidative stress that causes protein and DNA damage. The cell counters the stress response by increasing the expression of proteasomal degradation machinery. The cell also during the stress increases the activity of protein kinases (PKs) like ATM, ART, and DNA PKcs which activate CHK1 and CHK2 (DNA damage signaling proteins) and these then phosphorylate p53, which leads to apoptosis of the stressed cell. MMS-stressed cells also increase the expression of LC3 and P62/SQTM1 which leads to autophagy.
Future Directions
Given the intricate relationship between DNA damage, transcription regulation, and metabolism, future research should focus on:
-
Performing detailed functional studies of the identified proteins to elucidate their roles in the DDR. In addition, identifying the signaling pathways involved may help locate the specific action mechanism of MMS-induced carcinogenesis.
-
Investigating the potential therapeutic targets within these pathways to enhance cell survival or selectively induce death in cancer cells. Cancer cell lines and histologically confirmed cancerous tissues from different types of human malignancies may be evaluated to identify critical shifts in the metabolic proteome of the human system. Flow cytometry and microarrays may help identify the nature and cytotoxic potential of MMS toward different cell types, and targeting lymphocytes may help understand the immune status of the individual upon chronic MMS exposure.
-
Exploring the dynamic interactions between transcriptional and metabolic changes to develop a holistic understanding of cellular responses to DNA damage. Enzyme-linked immunosorbent assays and western blotting may be employed to check for any heightened immune responses as a result of accumulated DNA products or over-expression of protein products from different genes.
By advancing our knowledge in these areas, we can improve strategies for cancer treatment and enhance our ability to mitigate the adverse effects of genotoxic agents.
Conclusion
These findings highlight the complexity of cellular responses to DNA damage induced by MMS. The delay in the cell cycle, driven by changes in global transcription regulation, is accompanied by significant alterations in the proteome. Most of the available data discuss the methylation-induced genotoxicity of MMS and its potential to cause DNA damage directly or indirectly. This piece of literature review focused on MMS-induced OS, proteomic alterations, and metabolic shifts. The observed changes in the phosphorylation status of RNAP-II and the modulation of key metabolic pathways underscore the interconnectedness of DDR mechanisms and cellular metabolism. Further studies are warranted to explore the functional implications of these proteomic changes and understand how they contribute to cell survival or death following genotoxic stress.
This brief review article delved into the effects of the alkylating agent MMS on changes in the transcriptomic and proteomic pools of the cellular milieu, shedding light on the metabolic alterations as a result of MMS exposure. The findings can be extended to identify the MMS as a potential source of genome instability that can result in carcinogenic inductions and could also be associated with metabolic complications. Understanding the pathways and the specific enzymes affected could help identify the target sites for the molecular intervention. Ultimately, these observations could contribute to the broader knowledge of methylating agents. Extending these observations to the human cellular system and understanding the mechanisms of the cytotoxic impact of MMS will pave the way for future investigations and personalized therapeutic interventions, specifically in the area of metabolic diseases and cancers.
Moreover, at this stage, it can be inferred that MMS is not just a typical methylating agent; rather, it should be viewed as a toxicant capable of altering the genome, transcriptome, and metabolic proteome of the cellular system. The study presents a generalized aspect of MMS toxicity. Focusing specifically on human cell lines and cancers would help understand the situation better at the molecular level. We believe the study will open new avenues on the role of MMS in human carcinogenicity.
Authors’ Contribution
Resources: Moinuddin, Safia Habib.
Supervision: Safia Habib.
Visualization: Irfan Qadir Tantry, Safia Habib.
Writing–original draft: Mohd Mustafa.
Writing–review & editing: Badar ul Islam, Irfan Qadir Tantry, Moinuddin.
Competing Interests
The authors declare that they have no discernible competing financial interests or personal connections that could influence this work.
Ethical Approval
Not applicable.
Funding
This work is part of research supported by the Indian Council of Medical Research, New Delhi, India [Sanction Letter No. 45/24/2022-/ BIO/BMS] to Mohd Mustafa and the University Grants Commission, New Delhi, India [Project ID No. 2021-13007] to Safia Habib.
References
- Lipsick J. A history of cancer research: carcinogens and mutagens. Cold Spring Harb Perspect Med 2021; 11(3):a035857. doi: 10.1101/cshperspect.a035857 [Crossref] [ Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res 2005; 33(12):3799-811. doi: 10.1093/nar/gki681 [Crossref] [ Google Scholar]
- Mustafa M, Ali A, Siddiqui SA, Mir AR, Kausar T, Nayeem SM. Biophysical characterization of structural and conformational changes in methylmethane sulfonate modified DNA leading to the frizzled backbone structure and strand breaks in DNA. J Biomol Struct Dyn 2022; 40(16):7598-611. doi: 10.1080/07391102.2021.1899051 [Crossref] [ Google Scholar]
- Mustafa M, Habib S, Imtiyaz K, Tufail N, Ahmad R, Hamim B. Characterization of structural, genotoxic, and immunological effects of methyl methanesulfonate (MMS) induced DNA modifications: implications for inflammation-driven carcinogenesis. Int J Biol Macromol 2024; 268(Pt 1):131743. doi: 10.1016/j.ijbiomac.2024.131743 [Crossref] [ Google Scholar]
- Li S, Wang L, Wang Y, Zhang C, Hong Z, Han Z. The synthetic lethality of targeting cell cycle checkpoints and PARPs in cancer treatment. J Hematol Oncol 2022; 15(1):147. doi: 10.1186/s13045-022-01360-x [Crossref] [ Google Scholar]
- Bharati AP, Kumari S, Akhtar MS. Proteome analysis of Saccharomyces cerevisiae after methyl methane sulfonate (MMS) treatment. Biochem Biophys Rep 2020; 24:100820. doi: 10.1016/j.bbrep.2020.100820 [Crossref] [ Google Scholar]
- Feng Y, Zhang Y, Li J, Omran RP, Whiteway M, Feng J. Transcriptional profiling of the Candida albicans response to the DNA damage agent methyl methanesulfonate. Int J Mol Sci 2022; 23(14):7555. doi: 10.3390/ijms23147555 [Crossref] [ Google Scholar]
- Ji Y, Zhao M, Qiao X, Peng GH. Decitabine improves MMS-induced retinal photoreceptor cell damage by targeting DNMT3A and DNMT3B. Front Mol Neurosci 2022; 15:1057365. doi: 10.3389/fnmol.2022.1057365 [Crossref] [ Google Scholar]
- Araujo-Lima CF, Christoni LS, Justo G, Soeiro MN, Aiub CA, Felzenszwalb I. Atorvastatin downregulates in vitro methyl methanesulfonate and cyclophosphamide alkylation-mediated cellular and DNA injuries. Oxid Med Cell Longev 2018; 2018:7820890. doi: 10.1155/2018/7820890 [Crossref] [ Google Scholar]
- Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, Woodgate R. Eukaryotic Y-family polymerases bypass a 3-methyl-2’-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic Acids Res 2008; 36(7):2152-62. doi: 10.1093/nar/gkn058 [Crossref] [ Google Scholar]
- Stern HR, Sefcikova J, Chaparro VE, Beuning PJ. Mammalian DNA polymerase kappa activity and specificity. Molecules 2019; 24(15):2805. doi: 10.3390/molecules24152805 [Crossref] [ Google Scholar]
- Siler J, Xia B, Wong C, Kath M, Bi X. Cell cycle-dependent positive and negative functions of Fun30 chromatin remodeler in DNA damage response. DNA Repair (Amst) 2017; 50:61-70. doi: 10.1016/j.dnarep.2016.12.009 [Crossref] [ Google Scholar]
- Yao S, Feng Y, Zhang Y, Feng J. DNA damage checkpoint and repair: from the budding yeast Saccharomyces cerevisiae to the pathogenic fungus Candida albicans. Comput Struct Biotechnol J 2021; 19:6343-54. doi: 10.1016/j.csbj.2021.11.033 [Crossref] [ Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 2007; 318(5857):1780-2. doi: 10.1126/science.1145977 [Crossref] [ Google Scholar]
- Bharati AP, Singh N, Kumar V, Kashif M, Singh AK, Singh P. The mRNA capping enzyme of Saccharomyces cerevisiae has dual specificity to interact with CTD of RNA Polymerase II. Sci Rep 2016; 6:31294. doi: 10.1038/srep31294 [Crossref] [ Google Scholar]
- Jeong SJ, Kim HJ, Yang YJ, Seol JH, Jung BY, Han JW. Role of RNA polymerase II carboxy terminal domain phosphorylation in DNA damage response. J Microbiol 2005; 43(6):516-22. [ Google Scholar]
- Burger K, Ketley RF, Gullerova M. Beyond the trinity of ATM, ATR, and DNA-PK: multiple kinases shape the DNA damage response in concert with RNA metabolism. Front Mol Biosci 2019; 6:61. doi: 10.3389/fmolb.2019.00061 [Crossref] [ Google Scholar]
- Kim DR, Gidvani RD, Ingalls BP, Duncker BP, McConkey BJ. Differential chromatin proteomics of the MMS-induced DNA damage response in yeast. Proteome Sci 2011; 9:62. doi: 10.1186/1477-5956-9-62 [Crossref] [ Google Scholar]
- Kitanovic A, Walther T, Loret MO, Holzwarth J, Kitanovic I, Bonowski F. Metabolic response to MMS-mediated DNA damage in Saccharomyces cerevisiae is dependent on the glucose concentration in the medium. FEMS Yeast Res 2009; 9(4):535-51. doi: 10.1111/j.1567-1364.2009.00505.x [Crossref] [ Google Scholar]
- Kitanovic A, Wölfl S. Fructose-1,6-bisphosphatase mediates cellular responses to DNA damage and aging in Saccharomyces cerevisiae. Mutat Res 2006; 594(1-2):135-47. doi: 10.1016/j.mrfmmm.2005.08.005 [Crossref] [ Google Scholar]
- Blaszczak E, Pasquier E, Le Dez G, Odrzywolski A, Lazarewicz N, Brossard A. Dissecting ubiquitylation and DNA damage response pathways in the yeast Saccharomyces cerevisiae using a proteome-wide approach. Mol Cell Proteomics 2024; 23(1):100695. doi: 10.1016/j.mcpro.2023.100695 [Crossref] [ Google Scholar]
- Rochette S, Gagnon-Arsenault I, Diss G, Landry CR. Modulation of the yeast protein interactome in response to DNA damage. J Proteomics 2014; 100:25-36. doi: 10.1016/j.jprot.2013.11.007 [Crossref] [ Google Scholar]
- Li J, Eu JQ, Kong LR, Wang L, Lim YC, Goh BC. Targeting metabolism in cancer cells and the tumour microenvironment for cancer therapy. Molecules 2020; 25(20):4831. doi: 10.3390/molecules25204831 [Crossref] [ Google Scholar]
- Huang D, Piening BD, Kennedy JJ, Lin C, Jones-Weinert CW, Yan P. DNA replication stress phosphoproteome profiles reveal novel functional phosphorylation sites on Xrs2 in Saccharomyces cerevisiae. Genetics 2016; 203(1):353-68. doi: 10.1534/genetics.115.185231 [Crossref] [ Google Scholar]
- Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 2009; 34(2):168-78. doi: 10.1016/j.molcel.2009.02.025 [Crossref] [ Google Scholar]
- Czeko E, Seizl M, Augsberger C, Mielke T, Cramer P. Iwr1 directs RNA polymerase II nuclear import. Mol Cell 2011; 42(2):261-6. doi: 10.1016/j.molcel.2011.02.033 [Crossref] [ Google Scholar]
- Kim DR, Gidvani RD, Ingalls BP, Duncker BP, McConkey BJ. Differential chromatin proteomics of the MMS-induced DNA damage response in yeast. Proteome Sci 2011; 9:62. doi: 10.1186/1477-5956-9-62 [Crossref] [ Google Scholar]
- Davalli P, Marverti G, Lauriola A, D’Arca D. Targeting oxidatively induced DNA damage response in cancer: opportunities for novel cancer therapies. Oxid Med Cell Longev 2018; 2018:2389523. doi: 10.1155/2018/2389523 [Crossref] [ Google Scholar]
- Truman AW, Kristjansdottir K, Wolfgeher D, Ricco N, Mayampurath A, Volchenboum SL. Quantitative proteomics of the yeast Hsp70/Hsp90 interactomes during DNA damage reveal chaperone-dependent regulation of ribonucleotide reductase. J Proteomics 2015; 112:285-300. doi: 10.1016/j.jprot.2014.09.028 [Crossref] [ Google Scholar]
- Kumar A, Mathew V, Stirling PC. Dynamics of DNA damage-induced nuclear inclusions are regulated by SUMOylation of Btn2. Nat Commun 2024; 15(1):3215. doi: 10.1038/s41467-024-47615-8 [Crossref] [ Google Scholar]
- Pehlivan Ö C, Cavuşoğlu K, Yalçin E, Acar A. In silico interactions and deep neural network modeling for toxicity profile of methyl methanesulfonate. Environ Sci Pollut Res Int 2023; 30(55):117952-69. doi: 10.1007/s11356-023-30465-0 [Crossref] [ Google Scholar]
- Kitanovic A, Walther T, Loret MO, Holzwarth J, Kitanovic I, Bonowski F. Metabolic response to MMS-mediated DNA damage in Saccharomyces cerevisiae is dependent on the glucose concentration in the medium. FEMS Yeast Res 2009; 9(4):535-51. doi: 10.1111/j.1567-1364.2009.00505.x [Crossref] [ Google Scholar]
- Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 2020; 11(1):102. doi: 10.1038/s41467-019-13668-3 [Crossref] [ Google Scholar]
- Carter RJ, Parsons JL. Base excision repair, a pathway regulated by posttranslational modifications. Mol Cell Biol 2016; 36(10):1426-37. doi: 10.1128/mcb.00030-16 [Crossref] [ Google Scholar]
- Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen 2017; 58(5):235-63. doi: 10.1002/em.22087 [Crossref] [ Google Scholar]
- Bobyk L, Vianna F, Martinez JS, Gruel G, Benderitter M, Baldeyron C. Differential recruitment of DNA repair proteins KU70/80 and RAD51 upon microbeam irradiation with α-particles. Biology (Basel) 2022; 11(11):1652. doi: 10.3390/biology11111652 [Crossref] [ Google Scholar]
- Song HY, Shen R, Mahasin H, Guo YN, Wang DG. DNA replication: mechanisms and therapeutic interventions for diseases. MedComm (2020) 2023; 4(1):e210. doi: 10.1002/mco2.210 [Crossref] [ Google Scholar]
- Guo J, Yi GZ, Liu Z, Sun X, Yang R, Guo M. Quantitative proteomics analysis reveals nuclear perturbation in human glioma U87 cells treated with temozolomide. Cell Biochem Funct 2020; 38(2):185-94. doi: 10.1002/cbf.3459 [Crossref] [ Google Scholar]
- Li W, Li F, Zhang X, Lin HK, Xu C. Insights into the post-translational modification and its emerging role in shaping the tumor microenvironment. Signal Transduct Target Ther 2021; 6(1):422. doi: 10.1038/s41392-021-00825-8 [Crossref] [ Google Scholar]
- Podhorecka M, Skladanowski A, Bozko P. H2AX phosphorylation: its role in DNA damage response and cancer therapy. J Nucleic Acids 2010;2010. 10.4061/2010/920161.
- Ding L, Cao J, Lin W, Chen H, Xiong X, Ao H. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci 2020; 21(6):1960. doi: 10.3390/ijms21061960 [Crossref] [ Google Scholar]
- Milo-Cochavi S, Pareek M, Delulio G, Almog Y, Anand G, Ma LJ. The response to the DNA damaging agent methyl methanesulfonate in a fungal plant pathogen. Fungal Biol 2019; 123(5):408-22. doi: 10.1016/j.funbio.2019.03.007 [Crossref] [ Google Scholar]
- Wang H, Guo M, Wei H, Chen Y. Targeting p53 pathways: mechanisms, structures, and advances in therapy. Signal Transduct Target Ther 2023; 8(1):92. doi: 10.1038/s41392-023-01347-1 [Crossref] [ Google Scholar]
- Shi G, Zhou X, Wang X, Zhang X, Zhang P, Feng S. Signatures of altered DNA methylation gene expression after central and peripheral nerve injury. J Cell Physiol 2020; 235(6):5171-81. doi: 10.1002/jcp.29393 [Crossref] [ Google Scholar]